The University of Liverpool (UoL) wanted to understand and map the chemical space of small molecules. The chemical space has been estimated to amount to some 10 decillion (1060) molecules, although the largest current database covers approximately 11 billion examples. To achieve this, researchers used transformers, however, these are very memory hungry due to the quadratic dependence on string size of the standard version. UoL’s previous system, equipped with NVIDIA V100 GPUs was limited to about 150,000 molecules, but the team needed more compute power to better understand and identify molecular structure.
With funding from the UK Research and Innovation (UKRI) Biotechnology and Biological Sciences Research Council (BBSRC), UoL deployed a new NVIDIA DGX A100 system to get the processing power needed. To learn the relationship between mass spectra and molecular structure, UoL trained transformers with around seven million molecules. With augmented data, the data set was increased to 21 million molecules. Using this data, UoL were able to find the first solution for the structure identification problem of molecules not in existing databases -- a real breakthrough. With this system, they were able to increase the number of molecules to around six million, along with a significant increase in the rate of learning.
Project Approach
The UoL team primarily used the Ampere-based GPUs within their NVIDIA DGX systems to train four main deep learning models, which together contribute in the increase in understanding of the molecular space.
DeepGraphMolGen
With a Graph Convolutional Neural Network (GCN) trained using the policies of reinforcement learning, the team were able to develop, predict or generate molecules with desirable properties. The environment could lend, score or reward by evaluating each molecule predicted by GCN, with the highest rewards corresponding to the molecules with the most desirable properties. This meant the GCN was subsequently able to learn to predict molecules with the highest rewards attached.
VAE-Sim
The team at UoL created a Variational Autoencoder Network (VAE) as a novel approach to estimate the essential similarity between molecules. The bow-tie shaped network is able to generate the latent representation of every molecule, passed through a simplified molecular-input line-entry system (SMILES). The VAE model was trained to then maximise the similarity between the molecules that were actually similar.
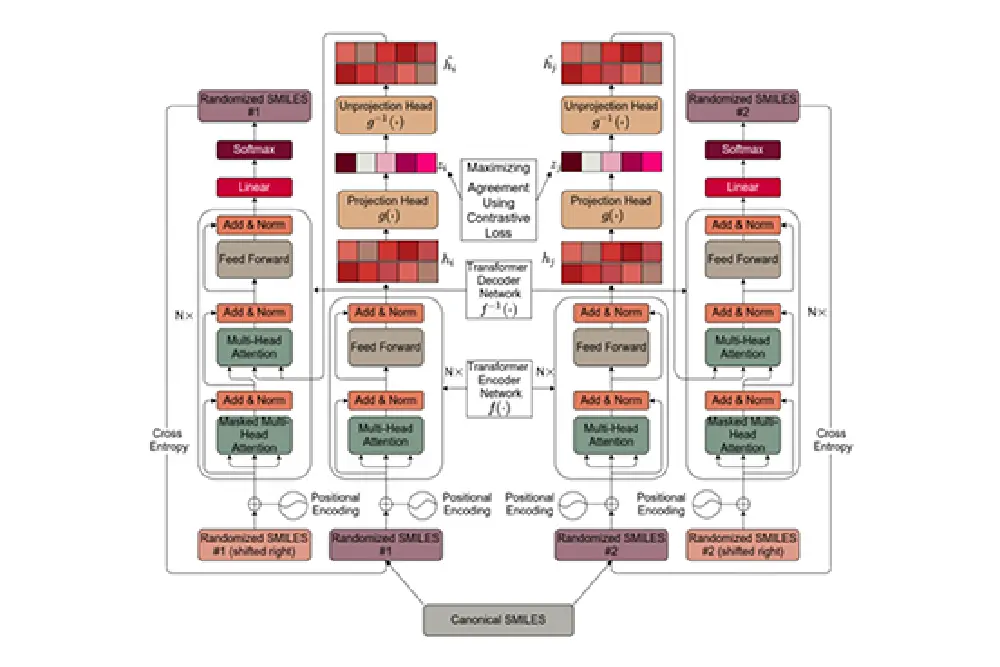
FragNet
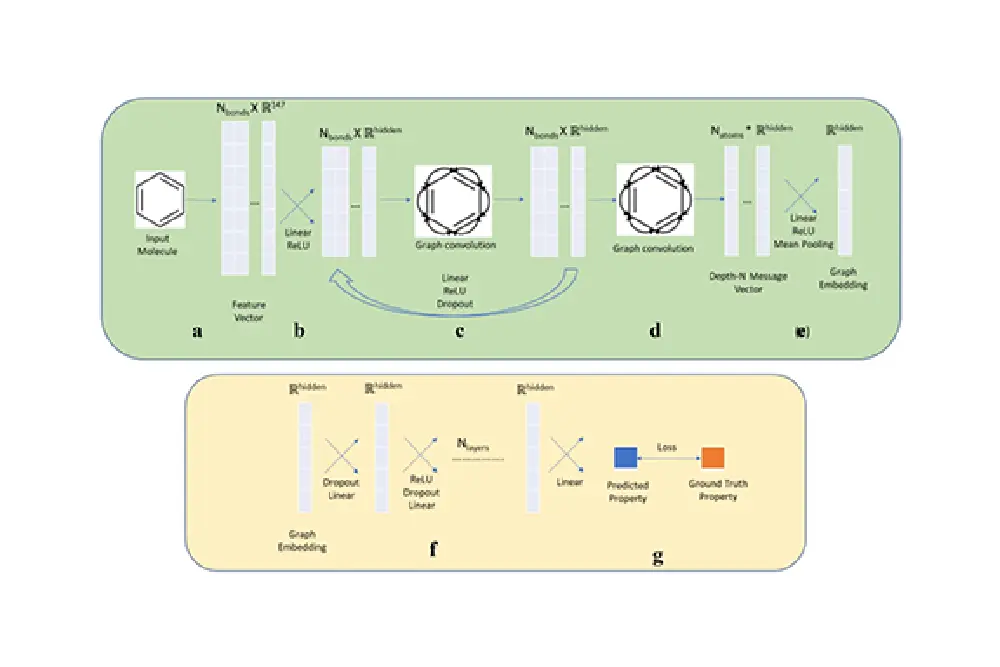
Next, the UoL team developed a novel architecture combining elements of transformers, auto-encoders and contrastive learning. The hybrid of transformers and auto-encoders were designed to predict the embedding of molecules, and contrastive learning was trained to have similar embedding for the molecules that were very similar. This resulted in a large, multi-dimensional latent space where similar molecules were clustered together and dissimilar molecules were far apart.
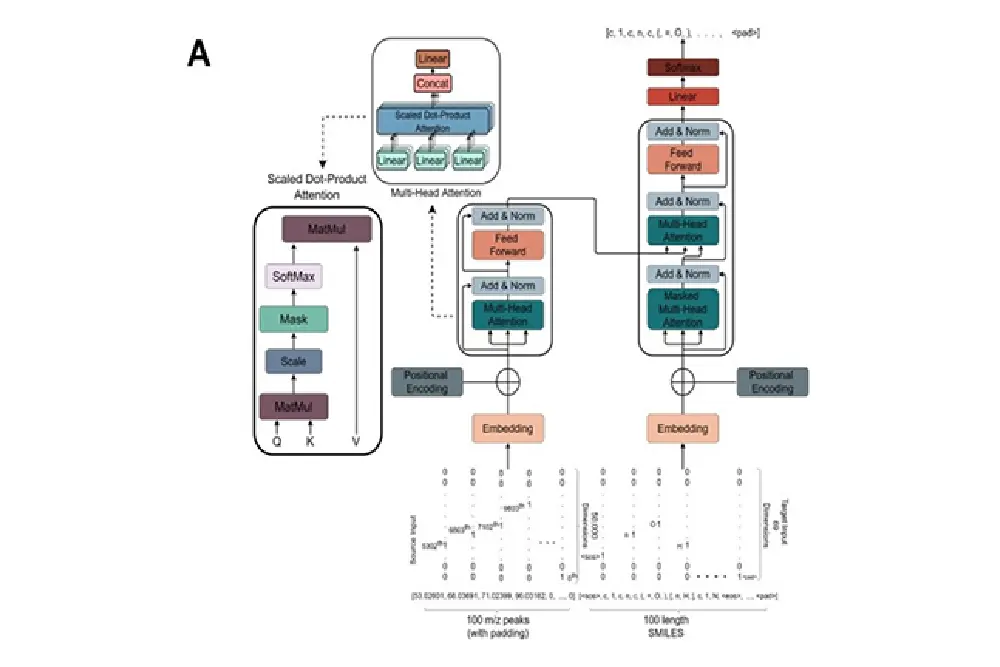
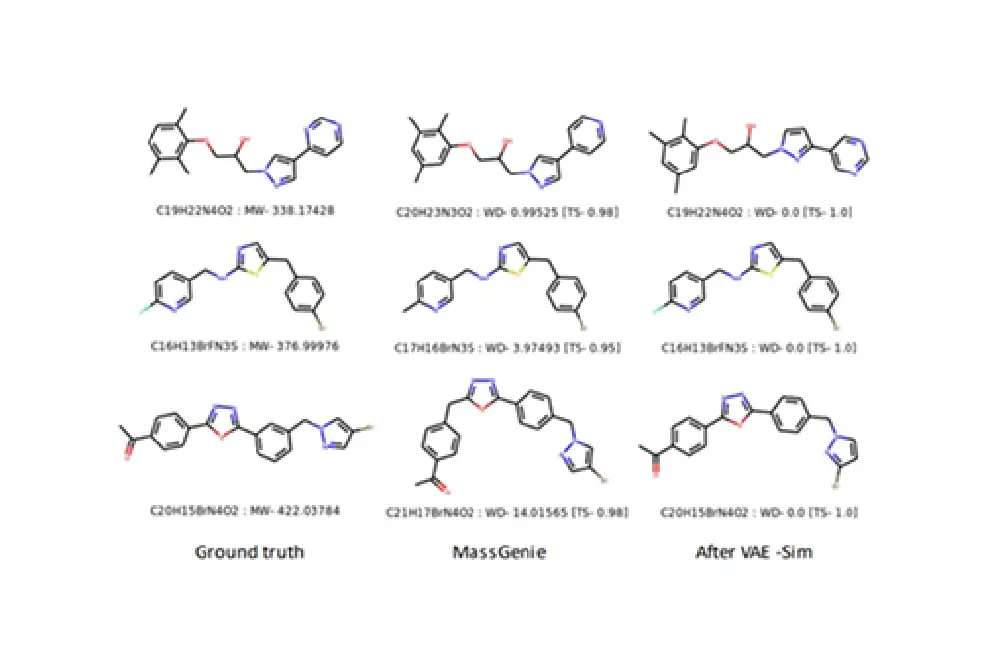
MassGenie
In this project, the team of researchers trained a large transformer-based deep neural network with around 6 million chemical structures to predict a SMILES representation of the molecules from their protonated mass spectra. MassGenie can learn the effective properties of the mass spectral fragment and valency space, and can generate candidate molecular structures that are very close or identical to those of the ‘true’ molecules.
With a Graph Convolutional Neural Network (GCN) trained using the policies of reinforcement learning, the team were able to develop, predict or generate molecules with desirable properties. The environment could lend, score or reward by evaluating each molecule predicted by GCN, with the highest rewards corresponding to the molecules with the most desirable properties. This meant the GCN was subsequently able to learn to predict molecules with the highest rewards attached.
Next Steps: Further Increasing the Chemical Space
The transformers trained for MassGenie used just a simple variety, which suffer from the well-known problem of a quadratic dependence on string length. To get round this issue, many newer versions have been proposed in terms of both algorithms and architectures. Changing the system build means the training set and transformer can be substantially increased, which increases the chemical space covered. UoL’s original work only covered mass spectra created using positive ionisation. For their next phase of research, the team will extend the approach to negative electrospray mass spectra.
The Scan Partnership
NVIDIA is a key partner of the University of Liverpool, and Scan was asked to act as a trusted advisor to help design, install and configure a DGX A100 infrastructure to aid the acceleration and scale of the research. The DGX A100 server was accompanied by NVIDIA Networking switches and connected to an AI-optimised PNY 3S-2450 storage appliance. NVIDIA and Scan were also on hand throughout the research to ensure maximum performance of the CUDA software and server and storage hardware.
Project Wins
40x increase in scope of molecular analysis from 150,000 to ~6 million
Up to 30x increase in speed of learning of molecular structure.
Better detection of molecular similarities, mass spectral fragment and valency space

Professor Douglas Kell
Research Chair in Systems Biology, University of Liverpool
"With our NVIDIA DGX A100 solution, we were able to increase our molecule analysis some 40-fold, along with a speed up in learning of between 10-30 fold"
Speak to an expert
You've seen how Scan has helped the University of Liverpool to accelerate molecule analysis. Contact our expert team to discuss your project requirements.
phone_iphone Phone: 01204 474210
mail Email: [email protected]